Cosmo Tutorial#
This is a brief guide on how to use GWFish to produce cosmological inferences
#! pip install -q git+https://github.com/janosch314/GWFish.git
#! pip install -q lalsuite
#! pip install -q corner
#! pip install -q dill
#! pip install -q getdist
Import packages#
# suppress warning outputs for using lal in jupuyter notebook
import warnings
warnings.filterwarnings("ignore", "Wswiglal-redir-stdio")
import GWFish.modules as gw
from tqdm import tqdm
import matplotlib
import matplotlib.pyplot as plt
import corner
import numpy as np
import pandas as pd
import json
import os
import dill as pickle
from getdist import loadMCSamples
import getdist.plots as gdplt
from scipy.integrate import simpson
import astropy.cosmology as apcosmo
from scipy.interpolate import interp1d
# plotting settings
matplotlib.rcParams.update({
'font.size': 10,
'axes.labelsize': 10,
'axes.titlesize': 16,
'xtick.labelsize': 10,
'ytick.labelsize': 10,
'legend.fontsize': 10,
'figure.titlesize': 14
})
Let’s create suitable catalog of events#
from_deg_to_rad = np.pi/180.
def create_dir(path):
if not os.path.exists(path):
os.makedirs(path)
print('Directory created: ', path)
else:
pass
#print('Directory already exists: ', path)
ns = 80
np.random.seed(42)
unphysical approach#
# data = {}
# data['redshift'] = np.random.uniform(0.001, 3, ns)
# mass_1_source = np.random.uniform(low = 1.1, high = 2.1, size = ns)
# mass_2_source = np.random.uniform(low = 1.1, high = 2.1, size = ns)
# data['luminosity_distance'] = np.ones(len(data['redshift']))#dL_z(data['redshift'])
# data['ra'] = np.random.uniform(0., 2 * np.pi, ns)
# data['dec'] = np.arcsin(np.random.uniform(-1., 1., ns))
# data['psi'] = np.random.uniform(0., np.pi, ns)
# data['phase'] = np.random.uniform(0., 2 * np.pi, ns)
# data['geocent_time'] = np.random.uniform(1577491218, 1609027217, ns)
physical approach#
def md_merger_rate(z, alphaF, betaF, CF, norm):
return norm * (1+z)**alphaF / (1+((1+z)/CF)**betaF)
def sampling_redshift_bns(redshift_dict, NSamples, seed):
#choosing the cosmology
keys = list(redshift_dict.keys())
if 'Om0' in keys:
Om0 = redshift_dict['Om0']
else:
Om0 = None
if 'Ode0' in keys:
Ode0 = redshift_dict['Ode0']
else:
Ode0 = None
if 'H0' in keys:
H0 = redshift_dict['H0']
else:
H0 = None
if None in (Om0,Ode0,H0):
cosmo = apcosmo.Planck18
else:
cosmo = apcosmo.LambdaCDM(H0=H0, Om0=Om0, Ode0=Ode0)
# Initializing the random number generator
rng = np.random.default_rng(seed)
# uniform redshift distribution
if redshift_dict['model'] == 'uniform':
if redshift_dict['default'] == True:
zmin = 0.
zmax = 10.
else:
zmin = redshift_dict['zmin']
zmax = redshift_dict['zmax']
z_vec = rng.uniform(low=zmin, high=zmax, size=NSamples)
# uniform in comoving volume
elif redshift_dict['model'] == 'uniform_comoving_volume':
if redshift_dict['default'] == True:
zmin = 0.
zmax = 10.
else:
zmin = redshift_dict['zmin']
zmax = redshift_dict['zmax']
nzs = max(5001, 50 * int((zmax-zmin)) + 1)
zs = np.linspace(zmin,zmax,nzs)
z_dist = (lambda z: ((4.*np.pi*cosmo.differential_comoving_volume(z).value)/(1.+z)))(zs)
window_cdf = np.array([simpson(z_dist[:i],zs[:i]) for i in range(1,nzs+1)]) / simpson(z_dist,zs)
inv_window_cdf = interp1d(window_cdf, zs)
z_vec = inv_window_cdf(rng.random(NSamples))
# Madau-Dickinson merger rate, with fiducial parameters come from the GWBENCH package
elif redshift_dict['model'] == 'madau_dickinson':
if redshift_dict['default'] == True:
zmin = 0.
zmax = 10.
alphaF = 1.803219571
betaF = 5.309821767
CF = 2.837264101
norm = 8.765949529
else:
zmin = redshift_dict['zmin']
zmax = redshift_dict['zmax']
alphaF = redshift_dict['alphaF']
betaF = redshift_dict['betaF']
CF = redshift_dict['CF']
norm = redshift_dict['norm']
nzs = max(5001, 50 * int((zmax-zmin)) + 1)
zs = np.linspace(zmin,zmax,nzs)
z_dist = (lambda z: ((md_merger_rate(z,alphaF,betaF,CF,norm)*4.*np.pi*cosmo.differential_comoving_volume(z).value)/(1.+z)))(zs)
window_cdf = np.array([simpson(z_dist[:i],zs[:i]) for i in range(1,nzs+1)]) / simpson(z_dist,zs)
inv_window_cdf = interp1d(window_cdf, zs)
z_vec = inv_window_cdf(rng.random(NSamples))
DL_vec = cosmo.luminosity_distance(z_vec).value
return z_vec, DL_vec
redshift_dict = {
'model' : 'madau_dickinson',
'default' : True
}
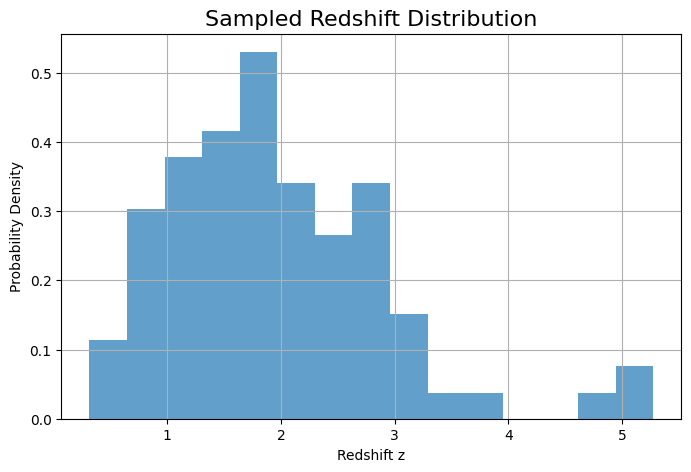
z_samples, dL_samples = sampling_redshift_bns(redshift_dict, ns, seed=42)
#plot z_samples
plt.figure(figsize=(8,5))
plt.hist(z_samples, bins=15, density=True, alpha=0.7)
plt.xlabel('Redshift z')
plt.ylabel('Probability Density')
plt.title('Sampled Redshift Distribution')
plt.grid()
plt.show()
# sort the two masses so that m1>m2 and create M_chirp and q
mass_1_source = np.random.uniform(low = 1.1, high = 2.1, size = ns)
mass_2_source = np.random.uniform(low = 1.1, high = 2.1, size = ns)
mass_1_det = mass_1_source * (1+z_samples)
mass_2_det = mass_2_source * (1+z_samples)
aux_mass = mass_1_det
mass_1_det = np.maximum(aux_mass, mass_2_det)
mass_2_det = np.minimum(aux_mass, mass_2_det)
#create only visible GRBs
a, b, c, d = -1., np.cos(175/180*np.pi), np.cos(5/180*np.pi), 1.
# Specify relative probabilities
prob = np.array([b-a, d-c])
prob = prob/prob.sum() # Normalize to sum up to one
r = []
for ii in range(ns):
r.append(np.random.choice([np.arccos(np.random.uniform(a, b)), np.arccos(np.random.uniform(c, d))], p=prob))
theta_data = np.array(r)
data = {}
data['redshift'] = z_samples
data['mass_ratio'] = mass_2_det/mass_1_det
data['chirp_mass'] = (mass_1_det * mass_2_det)**(3/5) / (mass_1_det + mass_2_det)**(1/5)
data['luminosity_distance'] = dL_samples
data['ra'] = np.random.uniform(0., 2 * np.pi, ns)
data['dec'] = np.arcsin(np.random.uniform(-1., 1., ns))
data['theta_jn'] = theta_data
data['psi'] = np.random.uniform(0., np.pi, ns)
data['phase'] = np.random.uniform(0., 2 * np.pi, ns)
data['geocent_time'] = np.random.uniform(1577491218, 1609027217, ns)
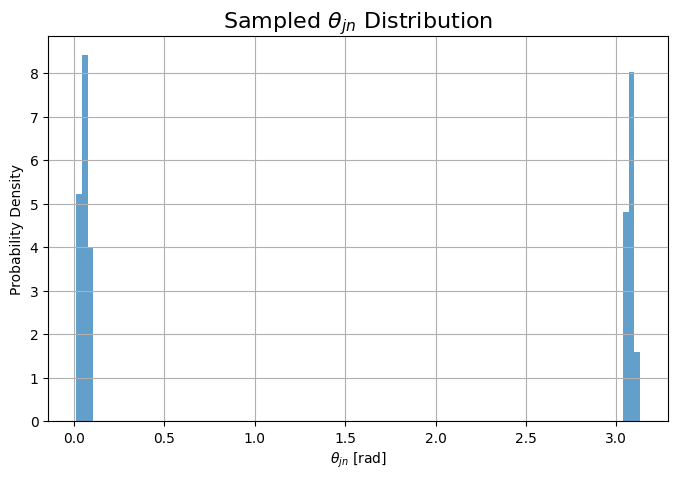
# plot theta_jn
plt.figure(figsize=(8,5))
plt.hist(data['theta_jn'], bins=100, density=True, alpha=0.7)
plt.xlabel(r'$\theta_{jn}$ [rad]')
plt.ylabel('Probability Density')
plt.title(r'Sampled $\theta_{jn}$ Distribution')
plt.grid()
plt.show()
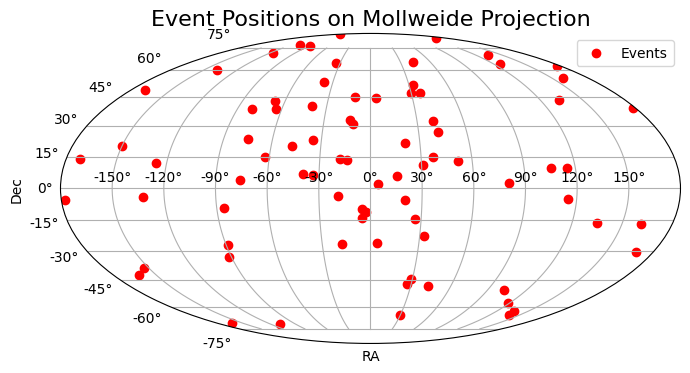
plt.figure(figsize=(8, 5))
ra_rad = data['ra'].astype(float)
# Shift RA from [0, 2pi] to [-pi, pi] for mollweide
ra_rad = np.where(ra_rad > np.pi, ra_rad - 2 * np.pi, ra_rad)
dec_rad = data['dec'].astype(float)
ax = plt.subplot(111, projection='mollweide')
ax.scatter(ra_rad, dec_rad, marker='o', color='red', label='Events')
ax.grid(True)
ax.set_xlabel('RA')
ax.set_ylabel('Dec')
ax.set_title('Event Positions on Mollweide Projection')
plt.legend()
plt.show()
data2save = pd.DataFrame(data=data)
data2save.head(5)
| redshift | mass_ratio | chirp_mass | luminosity_distance | ra | dec | theta_jn | psi | phase | geocent_time | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2.649630 | 0.751127 | 5.394567 | 22404.316540 | 0.647946 | 0.261777 | 0.052906 | 2.211932 | 0.126111 | 1.607083e+09 |
| 1 | 1.641375 | 0.840340 | 4.319453 | 12502.973009 | 5.670907 | 0.402258 | 0.083231 | 0.669047 | 2.023683 | 1.583207e+09 |
| 2 | 3.149753 | 0.781060 | 5.840114 | 27580.417558 | 3.174594 | -0.091044 | 3.092178 | 0.428424 | 1.328567 | 1.579588e+09 |
| 3 | 2.341390 | 0.684987 | 4.074958 | 19290.559269 | 5.192785 | 0.257968 | 3.074520 | 0.045693 | 2.057727 | 1.600863e+09 |
| 4 | 0.802911 | 0.890173 | 2.088720 | 5185.199461 | 2.010931 | 0.169438 | 0.076745 | 1.101403 | 0.752488 | 1.595608e+09 |
catalog_folder = os.path.join(os.getcwd(), "output")
create_dir(catalog_folder)
data2save.to_hdf(catalog_folder + f'/grb_events.hdf5', mode='w', key='root')
Let’s use GWFish! We will first use raw GWFish and then apply physical priors#
np.random.seed(42)
population = 'BNS'
networks = '[[0]]'
detectors = ['ET']
networks_ids = json.loads(networks)
ConfigDet = ' '
threshold_SNR = (0., 8.)
waveform_model = 'IMRPhenomHM'
fisher_parameters = ['chirp_mass', 'mass_ratio', 'luminosity_distance', 'theta_jn', 'psi', 'phase']
corner_lbs = ['$\mathcal{M}_c$ $[M_{\odot}]$', '$q$', '$d_L$ [Mpc]', '$\iota$ [rad]', '$\Psi$ [rad]', '$\phi$ [rad]']
parameters = pd.read_hdf(catalog_folder + f'/grb_events.hdf5')
parameters.head(5)
| redshift | mass_ratio | chirp_mass | luminosity_distance | ra | dec | theta_jn | psi | phase | geocent_time | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2.649630 | 0.751127 | 5.394567 | 22404.316540 | 0.647946 | 0.261777 | 0.052906 | 2.211932 | 0.126111 | 1.607083e+09 |
| 1 | 1.641375 | 0.840340 | 4.319453 | 12502.973009 | 5.670907 | 0.402258 | 0.083231 | 0.669047 | 2.023683 | 1.583207e+09 |
| 2 | 3.149753 | 0.781060 | 5.840114 | 27580.417558 | 3.174594 | -0.091044 | 3.092178 | 0.428424 | 1.328567 | 1.579588e+09 |
| 3 | 2.341390 | 0.684987 | 4.074958 | 19290.559269 | 5.192785 | 0.257968 | 3.074520 | 0.045693 | 2.057727 | 1.600863e+09 |
| 4 | 0.802911 | 0.890173 | 2.088720 | 5185.199461 | 2.010931 | 0.169438 | 0.076745 | 1.101403 | 0.752488 | 1.595608e+09 |
kwargs_network = {'detector_ids': detectors}
path_out_GWFish = os.path.join(os.getcwd(), "output", "gwfish_results")
if threshold_SNR is not None:
kwargs_network['detection_SNR'] = threshold_SNR
if ConfigDet!= ' ':
kwargs_network['config'] = ConfigDet
network = gw.detection.Network(**kwargs_network)
gw.fishermatrix.analyze_and_save_to_txt(network = network,
parameter_values = parameters,
fisher_parameters = fisher_parameters,
sub_network_ids_list = networks_ids,
population_name = population,
waveform_model = waveform_model,
save_path = path_out_GWFish,
save_matrices = True,
use_duty_cycle = True)
100%|██████████| 80/80 [00:09<00:00, 8.09it/s]
Let’s apply physical priors#
# join detector entries with underscore
detectors_str = '_'.join(detectors)
det = detectors_str # alias used later in the notebook
file_path = path_out_GWFish + f'/Errors_{detectors_str}_{population}_SNR{int(threshold_SNR[1])}.txt'
# Read header line (strip leading '#' or '//' and commas) and load data
with open(file_path, 'r') as f:
header = f.readline().lstrip('#/ \t').replace(',', ' ')
cols = header.split()
means = pd.read_csv(file_path, names=cols, skiprows=1, delim_whitespace=True)
cov_path = os.path.join(path_out_GWFish, f'inv_fisher_matrices_{detectors_str}_{population}_SNR{int(threshold_SNR[1])}.npy')
cov_matrix = np.load(cov_path)
lbs_mns = fisher_parameters
z_thr = 0.13
check_z_thr = means[means['redshift'] < z_thr]
CornerPlot = False
plotPosteriors = False
DensityPlot = False
PlotPeculiar = False
# if check_z_thr is empty then skip
if check_z_thr.empty:
include_vp = False
else:
include_vp = True
Example of Applications#
See Figure 1 in Cozzumbo, et al. 2024
mns = means[lbs_mns]
NN = len(mns)
N = 100_000
zmax = max(means['redshift'])
m1min = 1.1
m2min = 1.1
m1max = 2.5 * (1+zmax)
m2max = 2.5 * (1+zmax)
event_list = []
z_list = []
density_list = []
skipped_events = []
posterior_list = []
dl_inject = []
Ddl_inject = []
save_density = None
min_array = np.array([0., 0.01, 0., 0., 0., 0. ])
max_array = np.array([100., 1., np.inf, np.pi, np.pi, 2*np.pi ])
#from GWFish.modules.priors import *
from priors_mine import *
from scipy.stats import norm
mother_dir = os.getcwd()
# for each event we proceed to the truncation and then to the prior evaluation and posterior sampling from a physically motivated prior-informed likelihood
for event in tqdm(np.arange(NN)):
mns_ev = mns.iloc[event].to_numpy()
cov_ev = cov_matrix[event, :, :]
rng = np.random.default_rng(seed=100)
fisher_samples = pd.DataFrame(rng.multivariate_normal(mns_ev, cov_ev, N), columns = fisher_parameters)
# TRUNCATED SAMPLING
try:
with warnings.catch_warnings():
warnings.simplefilter("error") # Convert warnings into exceptions
samples_truncated_mvn = get_truncated_likelihood_samples(
fisher_parameters, mns_ev, cov_ev, N,
min_array=min_array, max_array=max_array
)
except Warning as w: # Catch warnings
print(f"Warning in truncated evaluation: {w}")
skipped_events.append([means['redshift'][int(event)]])
continue # Skip to the next iteration
except Exception as e: # Catch other exceptions
print(f"Error in truncated evaluation: {e}")
skipped_events.append([means['redshift'][int(event)]])
continue
priors_dict = get_default_priors_dict(fisher_parameters)
# Chirp mass prior
# Luminosity distance prior
priors_dict['luminosity_distance']['prior_type'] = "uniform_in_comoving_volume_and_source_frame"
priors_dict['luminosity_distance']['lower_prior_bound'] = 10
priors_dict['luminosity_distance']['upper_prior_bound'] = 50000
priors_dict['chirp_mass']['prior_type'] ='uniform_in_component_masses_chirp_mass'
priors_dict['chirp_mass']['lower_prior_bound'] = ((m1min * m2min)**(3/5) / (m1min + m2min)**(1/5))
priors_dict['chirp_mass']['upper_prior_bound'] = ((m1max * m2max)**(3/5) / (m1max + m2max)**(1/5))
# theta_jn prior
priors_dict['theta_jn']['prior_type'] = "uniform_in_sine"
theta_jn_lowerbound = 0.
theta_jn_upperbound = 5.
if 0 < means['theta_jn'][event] < np.pi/2 :
priors_dict['theta_jn']['lower_prior_bound'] = theta_jn_lowerbound*np.pi/180.
else:
priors_dict['theta_jn']['lower_prior_bound'] = (180-theta_jn_upperbound)*np.pi/180
if 0 < means['theta_jn'][event] < np.pi/2 :
priors_dict['theta_jn']['upper_prior_bound'] = theta_jn_upperbound*np.pi/180.
else:
priors_dict['theta_jn']['upper_prior_bound'] = (180-theta_jn_lowerbound)*np.pi/180
# PRIOR EVALUATION and POSTERIOR SAMPLING
try:
samples_from_posterior = get_posteriors_samples(fisher_parameters, samples_truncated_mvn, N, priors_dict = priors_dict)
except Exception as e:
print('Error in the prior evaluation:', e)
skipped_events.append([means['redshift'][int(event)]])
continue
z_save = means['redshift'][int(event)]
if CornerPlot:
CORNER_KWARGS = dict(
bins = 50, # number of bins for histograms
smooth = 0.99, # smooths out contours.
plot_datapoints = False, # choose if you want datapoints
truth_color='red',
label_kwargs = dict(fontsize = 16), # font size for labels
show_titles = True, #choose if you want titles on top of densities.
title_kwargs = dict(fontsize = 12), # font size for title
plot_density = True,
title_quantiles = [0.16, 0.5, 0.84], # add quantiles to plot densities for 1d hist
levels = (1 - np.exp(-0.5), 1 - np.exp(-2), 1 - np.exp(-9 / 2.)), # 1, 2 and 3 sigma contours for 2d plots
fill_contours = True, #decide if you want to fill the contours
max_n_ticks = 3, # set a limit to ticks in the x-y axes.
title_fmt=".2f"
)
fig = corner.corner(fisher_samples, labels = corner_lbs, truths = mns_ev, color = 'black', alpha = 0.5, **CORNER_KWARGS)
old_fig = corner.corner(samples_truncated_mvn, labels = corner_lbs, truths = mns_ev, color = 'orange', alpha = 0.5, **CORNER_KWARGS, fig = fig)
new_fig = corner.corner(samples_from_posterior, labels = corner_lbs, truths = mns_ev, color = 'red', alpha = 0.5, **CORNER_KWARGS, fig = fig)
plot_dir = f"{mother_dir}/check_images/corner"
create_dir(plot_dir)
plt.savefig(plot_dir + f'/cornerplot_{event}.pdf',dpi=600, bbox_inches='tight', facecolor='white')
plt.close()
if plotPosteriors:
# Set font size for ticks
matplotlib.rc('xtick', labelsize=8)
matplotlib.rc('ytick', labelsize=8)
# Create subplots
fig, axs = plt.subplots(2, 3, figsize=(10, 10))
# Histogram plots
params = fisher_parameters
positions = [(0, 0), (0, 1), (0, 2), (1, 0), (1, 1), (1, 2)]
for param, pos in zip(params, positions):
counts, edges, _ = axs[pos].hist(samples_from_posterior[param].to_numpy(), bins=50)
axs[pos].set_xlabel(param, fontsize=8)
# Plot injected values
axs[pos].axvline(x=mns_ev[params.index(param)], color='red', label='injected')
# Plot new mean values
axs[pos].axvline(x=samples_from_posterior[param].mean(), color='orange', label='new mean')
# Plot max posterior values
mode_index = np.argmax(counts)
axs[pos].axvline(x=0.5 * (edges[mode_index] + edges[mode_index + 1]), color='lightgreen', label='max posterior')
# Adjust layout and add legend
plt.legend(bbox_to_anchor=(1., 0.5), loc='center left', fontsize=8)
plt.tight_layout()
# Save plot
target_dir = f"{mother_dir}/check_images/posteriors"
create_dir(target_dir)
plt.savefig(f'{target_dir}/GW_posteriors_{event}.pdf', dpi=600, bbox_inches='tight', facecolor='white')
# Close figure
plt.close()
# here we save the posterior samples for the luminosity distance only, to be used in the cosmological analysis
# we use getdist kde interpolation, that is fast and flexible
# SAVE ALL POSTERIORS
weights, edges, _ = plt.hist(samples_from_posterior['luminosity_distance'].to_numpy(), bins=100, density=True)
plt.close()
out_path_posteriors = f"{mother_dir}/output/posteriors"
create_dir(out_path_posteriors)
final_samples = np.zeros((3,len(samples_from_posterior['luminosity_distance'].to_numpy())))
final_samples[0, :] = 1
final_samples[2,:] = samples_from_posterior['luminosity_distance'].to_numpy()
# CREATE PARAMFILE
param_list = ['d_L']
paramfile = [param_list[ii] + '\t' + param_list[ii] for ii in range(len(param_list))]
np.savetxt(out_path_posteriors + f'/posteriors_tmp.paramnames', paramfile, fmt='%s')
np.savetxt(out_path_posteriors+ f'/posteriors_tmp.txt', np.transpose(final_samples), fmt='%i %.6e %.6e')
settings = {'smooth_scale_1D' : 0.3}
sample_getdist=gdplt.loadMCSamples(out_path_posteriors + f'/posteriors_tmp', settings=settings)
density = gdplt.MCSamples.get1DDensityGridData(sample_getdist, 0)
z_list.append(z_save)
density_list.append(density)
if DensityPlot:
plt.rc('font', size=25, family='serif')
plt.rc('text', usetex=True)
fig = plt.figure(figsize=(10, 10))
xx = np.linspace(min(samples_from_posterior['luminosity_distance'].to_numpy()), max(samples_from_posterior['luminosity_distance'].to_numpy()),300)
plt.hist(samples_from_posterior['luminosity_distance'].to_numpy(), bins=100, density=True, alpha=0.7)
plt.plot(xx, max(weights)*density(xx), lw=3, label= 'Interpolating density')
#plt.plot(xx, max(weights)*density2(xx), label= 'Interpolating density bw=0.3')
plt.legend()
plot_dir = f"{mother_dir}/check_images/density"
create_dir(plot_dir)
plt.tight_layout()
plt.xlabel(r'$z$')
plt.ylabel(r'$p(d_L)$')
plt.savefig(plot_dir + f'/densityplot_{z_save}_{event}.pdf',dpi=600, bbox_inches='tight', facecolor='white')
#plt.show()
plt.close()
# just to save the posteriors
posterior_list.append(samples_from_posterior['luminosity_distance'].to_numpy())
dl_inject.append(means['luminosity_distance'].to_numpy()[event])
Ddl_inject.append(means['err_luminosity_distance'].to_numpy()[event])
out_path_kde = f"{mother_dir}/output/kde"
create_dir(out_path_kde)
# if there are elements with z < z_thr, then we apply the correction
if include_vp:
print('Peculiar velocity correction on going')
print('Starting the peculiar velocity analysis\n')
# include vp correction
dt = {}
dt['z'] = z_list
dt['posterior'] = posterior_list
dt['kde'] = density_list
dt['dl'] = dl_inject
dt['Ddl'] = Ddl_inject
dataframe_data = pd.DataFrame(data=dt)
sorted_df = dataframe_data.sort_values(by=['z'])
idx_cycle = np.where(np.array(sorted_df['z']) <= z_thr)[0]
mean_vp = 300.
sigma_vp = 200.
if len(idx_cycle) != 0:
for idx, ii in enumerate(idx_cycle):
gw_samples = sorted_df['posterior'].to_numpy()[ii]
gw_density = sorted_df['kde'].to_numpy()[ii]
z_event = sorted_df['z'].to_numpy()[ii]
dl= sorted_df['dl'].to_numpy()[ii]
Ddl= sorted_df['Ddl'].to_numpy()[ii]
rng = np.random.RandomState(ii)
vp_rms = rng.normal(loc=mean_vp, scale=50)
sigma_vp = rng.normal(loc=sigma_vp, scale=50)
sigma_dL_vp = np.median(gw_samples) * sigma_vp / (vp_rms + c * z_event)
#first estimate of the peculiar velocity distribution impedence when combined by
#simple quadrature sum because we need dL_values to be big enough
peculiar_Ddl = np.sqrt((Ddl)**2 + sigma_dL_vp**2)
if dl-3*peculiar_Ddl <=0:
min_bound = 0
else:
min_bound = dl-3*peculiar_Ddl
dL_values = np.linspace(min_bound, dl+3*peculiar_Ddl, 10000)
# Generate a Gaussian distribution for the peculiar velocity part
v_p_distribution = norm(loc=np.median(gw_samples), scale=sigma_dL_vp)
# Calculate the PDF of the peculiar velocity part
pdf_vp = v_p_distribution.pdf(dL_values)
# Calculate the PDF of the combined distribution by convolving the PDFs
combined_pdf = np.convolve(pdf_vp, gw_density(dL_values), mode='same')
# Normalize the combined PDF
combined_pdf /= np.sum(combined_pdf)
histogram, bin_edges = np.histogram(dL_values, bins=10000, weights=combined_pdf)
combined_samples = np.random.choice((bin_edges[:-1] + bin_edges[1:]) / 2, size=1_000_000, p= np.abs(histogram) / np.sum(histogram))
samp = np.zeros((3,len(combined_samples)))
samp[0, :] = 1
samp[2,:] = combined_samples
np.savetxt(out_path_posteriors+ f'/posteriors_tmp.txt', np.transpose(samp), fmt='%i %.6e %.6e')
settings = {'smooth_scale_1D' :0.3}
getdist=gdplt.loadMCSamples(out_path_posteriors+ f'/posteriors_tmp', settings=settings)
density_final = gdplt.MCSamples.get1DDensityGridData(getdist, 0)
dataframe_data['kde'] = density_final
#Plot the combined distribution
if PlotPeculiar:
plt.figure(figsize=(10, 10))
plt.plot(dL_values, density_final(dL_values), label=r'Combined kde with Gaussian $v_{\mathrm{peculiar}}$', color='green')
plt.plot(dL_values, gw_density(dL_values), label='Old kde', color='red')
plt.xlabel(r'$d_L$')
plt.ylabel('Density')
plt.title(f'Event at $z$={z_event} | $z_t$={z_thr} $v_p$={vp_rms:.2f} $\sigma_v$={sigma_vp:.2f}')
plt.legend(loc='upper right')
plt.xlim(np.min(gw_samples), np.max(gw_samples))
plot_dir = f"{mother_dir}/check_images/peculiar_velocity"
create_dir(plot_dir)
plt.savefig(plot_dir + f'/peculiar_velocity_{idx}.pdf',dpi=600, bbox_inches='tight', facecolor='white')
#plt.show()
plt.close()
density_vp = list(dataframe_data['kde'])
save_density = [z_list, density_list, dl_inject, Ddl_inject, density_vp]
else:
save_density = [z_list, density_list, dl_inject, Ddl_inject]
cosmology_save = 'LCDM_Planck18'
create_dir(out_path_kde + f'/{det}_{cosmology_save}_fiducial')
with open(out_path_kde + f'/{det}_{cosmology_save}_fiducial/{det}_{cosmology_save}_fiducial.pkl', 'wb') as handle:
pickle.dump(save_density, handle, protocol=pickle.HIGHEST_PROTOCOL)
os.remove(out_path_posteriors + f'/posteriors_tmp.txt')
print(f'Max z seen: {max(z_list)}')
print('Skipped events [name, redshift]:', skipped_events)
Let’s play with cosmology!#
we generate a likelihood and we run a cosmological MCMC with cobaya#
this is interfaced with a Einstein-Boltzmann solver called CLASS#
#! pip install -q cobaya
#! pip install -q openmpi
#! pip install -q "mpi4py>=3"
import cobaya
from cobaya.run import run
from cobaya.yaml import yaml_load_file
import yaml
# installing CLASS
os.system(f"cobaya-install classy -p {mother_dir}/myclass")
[install] Installing external packages at '/Users/andreacozzumbo/GSSIprojects/acme_tutorials/cosmo_tutorial/myclass'
[install] The installation path has been written into the global config file: /Users/andreacozzumbo/Library/Application Support/cobaya/config.yaml
================================================================================
classy
================================================================================
[install] Checking if dependencies have already been installed...
[install] External dependencies for this component already installed.
[install] Doing nothing.
================================================================================
* Summary *
================================================================================
[install] All requested components' dependencies correctly installed at /Users/andreacozzumbo/GSSIprojects/acme_tutorials/cosmo_tutorial/myclass
0
cosmology = 'LCDM_Planck18'
cosmo_data_path = os.path.join(mother_dir, "output", "kde", f"{det}_{cosmology}_fiducial")
pt_path = os.path.join(mother_dir, "cosmo_MCMC")
class_path = os.path.join(mother_dir, "myclass", "code", "classy")
output_path_MCMC = os.path.join(mother_dir, "cosmo_MCMC", "chains", "test1", "test")
input_yaml_path = os.path.join(mother_dir, "cosmo_MCMC", "input", "test.yaml")
#input_file = f"{mother_dir}/cosmo_MCMC/input/input.yaml"
base_config = {
"theory": {
"classy": {
"path": class_path,
"ignore_obsolete": True
}
},
"likelihood": {
"likelihood.MMcosmology": {
"data_directory": cosmo_data_path,
"python_path": pt_path,
"gw_network": "ET",
"grb_dataset": "fiducial",
"fiducial_cosmology": "LCDM_Planck18",
"interpolation_method": "kde",
}
},
"params": {
"omega_b": {
"value": 0.0224,
"latex": r"\Omega_\mathrm{b} h^2"
},
"Omega_cdm": {
"prior": {"min": 0.05, "max": 0.7},
"ref": {"dist": "norm", "loc": 0.26, "scale": 0.01},
"proposal": 0.05,
"latex": r"\Omega_\mathrm{c}"
},
"H0": {
"prior": {"min": 20, "max": 100},
"ref": {"dist": "norm", "loc": 67, "scale": 0.5},
"proposal": 1.,
"latex": "H_\mathrm{0}"
},
"Omega_m": {"latex": r"\Omega_\mathrm{m}"},
"Omega_Lambda": {"latex": r"\Omega_\Lambda"}
},
"sampler": {
"mcmc": {
"max_tries": 10000000,
#"covmat": '/home/cozzumbo/AC/Theseus/covmat/covmat_all.covmat',
"Rminus1_stop": 0.01
}
},
"output": output_path_MCMC
}
with open(input_yaml_path, "w") as f:
yaml.dump(base_config, f, sort_keys=False)
input_file = input_yaml_path
os.system(f'mpirun -n 4 cobaya-run {input_file}')
from getdist import loadMCSamples
from getdist import plots
import matplotlib as mpl
mpl.rcParams['text.usetex'] = False
settings = {'ignore_rows' : 0.3, 'smooth_scale_2D' : 0.5, 'smooth_scale_1D' : 0.5, 'contours':[0.68, 0.95, 0.997]}
samples = loadMCSamples(f"{mother_dir}/cosmo_MCMC/chains/test1/test", settings=settings)
WARNING:root:outlier fraction 0.003188097768331562
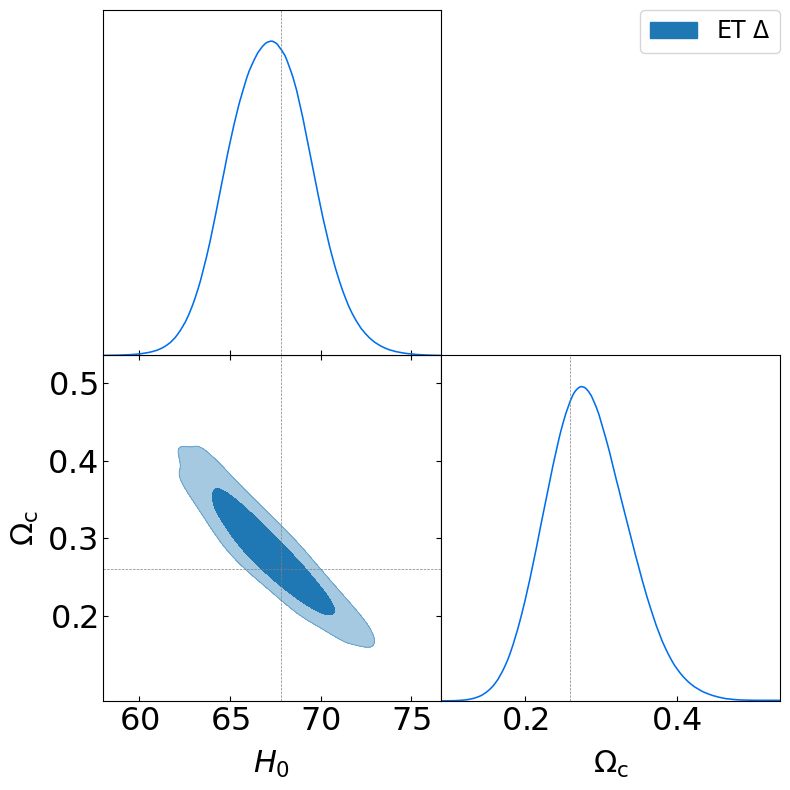
truth_LCDM={'Omega_m':0.309, 'Omega_cdm':0.26, 'H0':67.81}
g = plots.get_subplot_plotter()
g.settings.subplot_size_inch = 4
g.settings.legend_fontsize = 17
g.settings.axes_fontsize = 23
g.settings.lab_fontsize = 22
g.triangle_plot([samples],
params=['H0', 'Omega_cdm'] ,
filled=[True],
markers=truth_LCDM,
colors=['C0'],
legend_labels = [r"ET $\Delta$"],
legend_loc='upper right',
)